containerit packages R script/session/workspace and all dependencies as a Docker container by automagically generating a suitable Dockerfile. The package’s website is https://o2r.info/containerit/.
Prerequisites
-
containeritonly fully works if you have Docker installed and is only tested with Docker Engine - Community (previously called Docker Community Edition or Docker CE). -
R (>= 3.5.0)is needed so that some dependencies (e.g. BiocManager) are available; older versions of R predate the development of the package and were never tested.
Quick start
Try out containerit in a container
You can spin up a Docker container with containerit pre-installed if you want to try out the package. The default of the containerit images on Docker Hub is to start plain R, but you can also start an with RStudio session in a browser.
Note: The geospatial variant is used so that examples from containerit vignettes are likely to work. For a minimal Dockerfile to run containerit, see inst/docker/minimal/Dockerfile.
Now go to http://localhost:8787 and log in with the user rstudio and password o2r. Continue in section Use.
Use
containerit can create Dockerfile objects in R and render them as Dockerfile instructions based on session information objects or runnable R files (.R, .Rmd).
suppressPackageStartupMessages(library("containerit"))
my_dockerfile <- containerit::dockerfile(from = utils::sessionInfo())
#> INFO [2019-08-21 19:35:15] Going online? TRUE ... to retrieve system dependencies (sysreq-api)
#> INFO [2019-08-21 19:35:15] Trying to determine system requirements for the package(s) 'assertthat,backports,commonmark,crayon,curl,desc,digest,evaluate,formatR,fs,futile.logger,futile.options,htmltools,httpuv,jsonlite,knitr,lambda.r,later,magrittr,MASS,memoise,mime,miniUI,pillar,pkgconfig,pkgdown,promises,R6,Rcpp,rlang,rmarkdown,roxygen2,rprojroot,rstudioapi,semver,shiny,shinyFiles,stevedore,stringi,stringr,tibble,versions,xfun,xml2,xtable,yaml' from sysreqs online DB
#> INFO [2019-08-21 19:35:18] Adding CRAN packages: assertthat, backports, commonmark, crayon, curl, desc, digest, evaluate, formatR, fs, futile.logger, futile.options, htmltools, httpuv, jsonlite, knitr, lambda.r, later, magrittr, MASS, memoise, mime, miniUI, pillar, pkgconfig, pkgdown, promises, R6, Rcpp, rlang, rmarkdown, roxygen2, rprojroot, rstudioapi, semver, shiny, shinyFiles, stevedore, stringi, stringr, tibble, versions, xfun, xml2, xtable, yaml
#> INFO [2019-08-21 19:35:18] Created Dockerfile-Object based on sessionInfoprint(my_dockerfile)
#> FROM rocker/r-ver:3.6.1
#> LABEL maintainer="daniel"
#> RUN export DEBIAN_FRONTEND=noninteractive; apt-get -y update \
#> && apt-get install -y git-core \
#> libcurl4-openssl-dev \
#> libssl-dev \
#> libxml2-dev \
#> make \
#> pandoc \
#> pandoc-citeproc
#> RUN ["install2.r", "assertthat", "backports", "commonmark", "crayon", "curl", "desc", "digest", "evaluate", "formatR", "fs", "futile.logger", "futile.options", "htmltools", "httpuv", "jsonlite", "knitr", "lambda.r", "later", "magrittr", "MASS", "memoise", "mime", "miniUI", "pillar", "pkgconfig", "pkgdown", "promises", "R6", "Rcpp", "rlang", "rmarkdown", "roxygen2", "rprojroot", "rstudioapi", "semver", "shiny", "shinyFiles", "stevedore", "stringi", "stringr", "tibble", "versions", "xfun", "xml2", "xtable", "yaml"]
#> WORKDIR /payload/
#> CMD ["R"]You can disable logging:
Now we create a Dockerfile for a specific R version and R Markdown file and do not add any packages already available in the base image:
rmd_dockerfile <- containerit::dockerfile(from = "inst/demo.Rmd",
image = "rocker/verse:3.5.2",
maintainer = "o2r",
filter_baseimage_pkgs = TRUE)
#> Detected API version '1.40' is above max version '1.39'; downgrading
#> Detected API version '1.40' is above max version '1.39'; downgrading
print(rmd_dockerfile)
#> FROM rocker/verse:3.5.2
#> LABEL maintainer="o2r"
#> # CRAN packages skipped because they are in the base image: assertthat, backports, cli, crayon, curl, desc, digest, evaluate, formatR, fs, htmltools, httpuv, jsonlite, knitr, later, magrittr, mime, miniUI, pillar, pkgconfig, promises, R6, Rcpp, rlang, rmarkdown, rprojroot, rstudioapi, sessioninfo, shiny, stringi, stringr, tibble, withr, xfun, xtable, yaml
#> RUN ["install2.r", "fortunes", "futile.logger", "futile.options", "lambda.r", "semver", "shinyFiles", "stevedore", "versions"]
#> WORKDIR /payload/
#> CMD ["R"]For extended instructions, see the vignettes at in the directory vignettes/, which are readable online at https://o2r.info/containerit/articles/.
Images
Images are available starting from different base images. All images are also available with version tags.
The Dockerfiles are available in the directory inst/docker/Dockerfile.
RStudio Add-in
RStudio Addins allow to create interactive user interfaces for the RStudio development environment. Courtesy of a great contribution by a group of enthusiasts at the ROpenSci OZ Unconference 2018, there are several forms to quickly create Dockefiles from different use cases, e.g. the current session, a vector of expressions, or a script file.
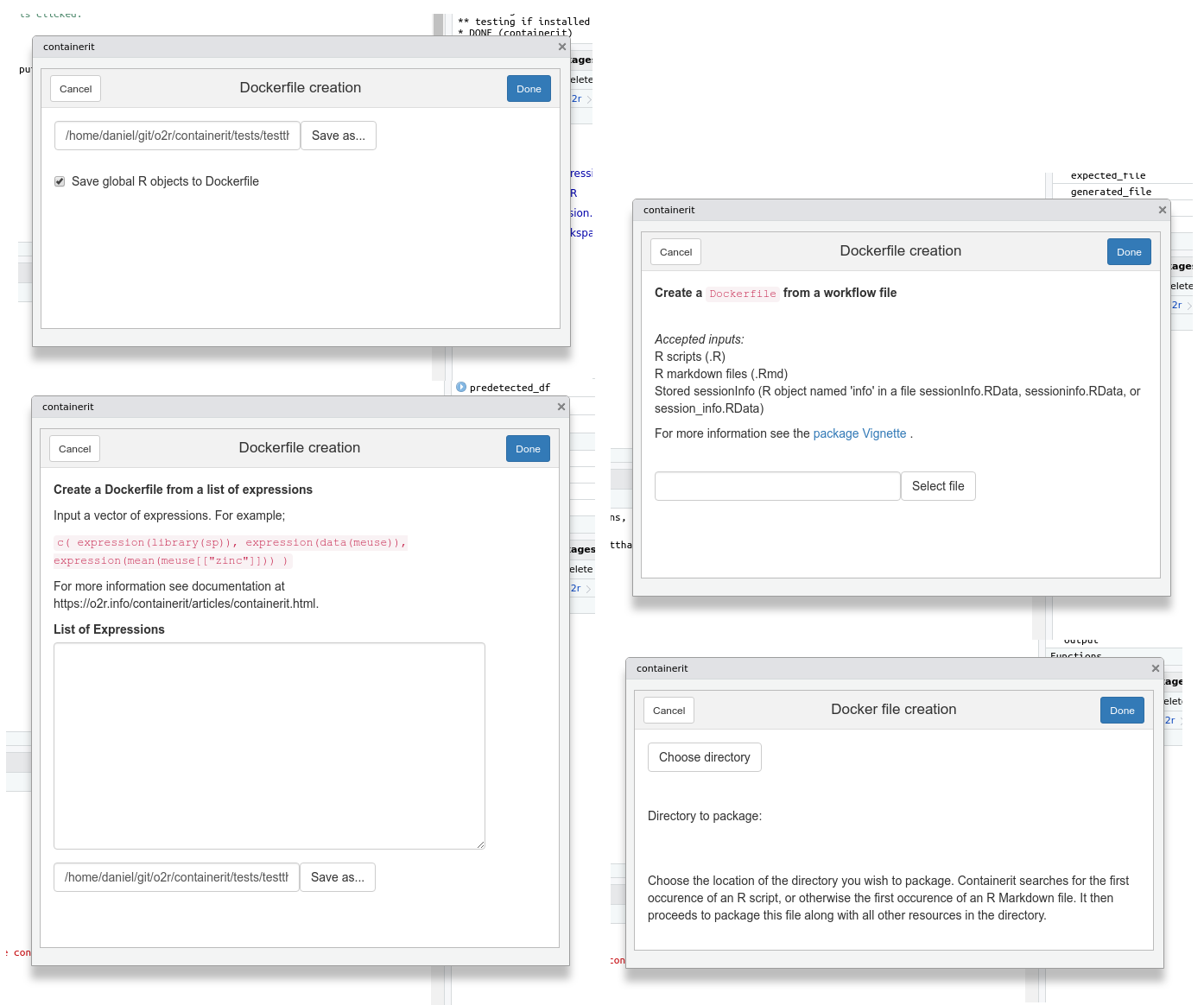
screenshots containerit RStudio Addin forms
Contribute
All help is welcome: asking questions, providing documentation, testing, or even development.
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
See CONTRIBUTING.md for details.
Development
r-hub builder is great for running checks, e.g. before submitting to CRAN and on other operating systems.
You can build the pkgdown site with
You can build the Docker images locally with the current development version using the following commands.
docker build --tag containerit:dev --file inst/docker/Dockerfile.local .
docker build --tag containerit:geospatial-dev --file inst/docker/geospatial/Dockerfile.local .You can use pre-commit hooks to avoid some mistakes.
A codemeta file, codemeta.json, with metadata about the package and its dependencies is generated automatically when this document is compiled.
Citation
citation("containerit")
#>
#> To cite containerit in publications use:
#>
#> Nüst, D. and Hinz, M. (2019). containerit: Generating
#> Dockerfiles for reproducible research with R. Journal of Open
#> Source Software, 4(40), 1603,
#> https://doi.org/10.21105/joss.01603
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {{containerit: Generating Dockerfiles for reproducible research with R}},
#> author = {Daniel Nüst and Matthias Hinz},
#> journal = {{Journal of Open Source Software}},
#> year = {2019},
#> month = {8},
#> volume = {4},
#> number = {40},
#> pages = {1603},
#> doi = {10.21105/joss.01603},
#> url = {https://doi.org/10.21105/joss.01603},
#> }





