How to increase reproducibility and transparency in your research
04 Feb 2019 | By Daniel Nüst[This article is cross posted-on the EGU GeoLog.]
Contemporary science faces many challenges in publishing results that are reproducible. This is due to increased usage of data and digital technologies as well as heightened demands for scholarly communication. These challenges have led to widespread calls for more research transparency, accessibility, and reproducibility from the science community. This article presents current findings and solutions to these problems, including recent new software that makes writing submission-ready manuscripts for journals of Copernicus Publications a lot easier. While it can be debated if science really faces a reproducibility crisis, the challenges of computer-based research have sparked numerous articles on new good research practices and their evaluation. The challenges have also driven researchers to develop infrastructure and tools to help scientists effectively write articles, publish data, share code for computations, and communicate their findings in a reproducible way, for example Jupyter, ReproZip and research compendia.
Recent studies showed that the geosciences and geographic information science are not beyond issues with reproducibility, just like other domains. Therefore, more and more journals have adopted policies on sharing data and code. However, it is equally important that scientists foster an open research culture and teach researchers how they adopt more transparent and reproducible workflows, for example at skill-building workshops at conferences offered by fellow researchers, such as the EGU short courses, community-led non-profit organisations such as the Carpentries, open courses for students, small discussion groups at research labs, or individual efforts of self-learning. In the light of prevailing issues of a common definition of reproducibility, Philip Stark, a statistics professor and associate dean of mathematical and physical sciences at the University of California, Berkeley, recently coined the term preproducibility: “An experiment or analysis is preproducible if it has been described in adequate detail for others to undertake it.” The neologism intends to reduce confusion and also to embrace a positive attitude for more openness, honesty, and helpfulness in scholarly communication processes.
In the spirit of these activities, this article describes a modern workflow made possible by recent software releases. The new features allow the EGU community to write preproducible manuscripts for submission to the large variety of academic journals published by Copernicus Publications. The new workflow might require hard-earned adjustments for some researchers, but it pays off because of an increase in transparency and effectivity. This is especially the case for early career scientists. An open and reproducible workflow enables researchers to build on others’ and own previous work and better collaborate on solving the societal challenges of today.
Reproducible research manuscripts
Open digital notebooks, which interweave data and code and can be exported to different output formats such as PDF, are powerful means to improve transparency and preproducibility of research. Jupyter Notebook, Stencila and R Markdown let researchers combine long-form text of a publication and source code for analysis and visualisation in a single document. Having text and code side-by-side makes them easier to grasp and ensures consistency, because each rendering of the document executes the whole workflow using the original data. Caching for long-lasting computations is possible, and researchers working with supercomputing infrastructures or huge datasets may limit the executed code to purposes of visualisation using processed data as input. Authors can transparently expose specific code snippets to readers but also publish the complete source code of the document openly for collaboration and review.
The popular notebook formats are plain text-based, like Markdown in case of R Markdown. Therefore an R Markdown document can be managed with version control software, which are programs for managing multiple versions and contributions, even by different people, to the same documents. Version control provides traceability of authorship, a time machine for going back to any previous “working” version, and online collaboration such as on GitLab. This kind of workflow also stops the madness of using file names for versions yet still lets authors use awesome file names and apply domain-specific guidelines for packaging research.
Final.doc https://t.co/YXJaSacHWu pic.twitter.com/4bBDzn7TXt
— PHD Comics (@PHDcomics) 1. Februar 2017
R Markdown supports different programming languages besides the popular namesake R and is a sensible solution even if you do not analyse data with scripts nor have any code in your scholarly manuscript. It is easy to write, allows you to manage your bibliography effectively, can be used for websites, books or blogs, but most importantly it does not fall short when it is time to submit a manuscript article to a journal.
The rticles extension package for R provides a number of templates for popular journals and publishers.
Since version 0.6 (published Oct 9 2018) these templates include the Copernicus Publications Manuscript preparations guidelines for authors.
The Copernicus Publications staff was kind enough to give a test document a quick review and all seems in order, though of course any problems and questions shall be directed to the software’s vibrant community and not the publishers.
The following code snippet and screen shot demonstrate the workflow.
Lines starting with # are code comments and explain the steps.
Code examples provided here are ready to use and only lack the installation commands for required packages.
# load required R extension packages:
library("rticles")
library("rmarkdown")
# create a new document using a template:
rmarkdown::draft(file = "MyArticle.Rmd",
template = "copernicus_article",
package = "rticles", edit = FALSE)
# render the source of the document to the default output format:
rmarkdown::render(input = "MyArticle/MyArticle.Rmd")
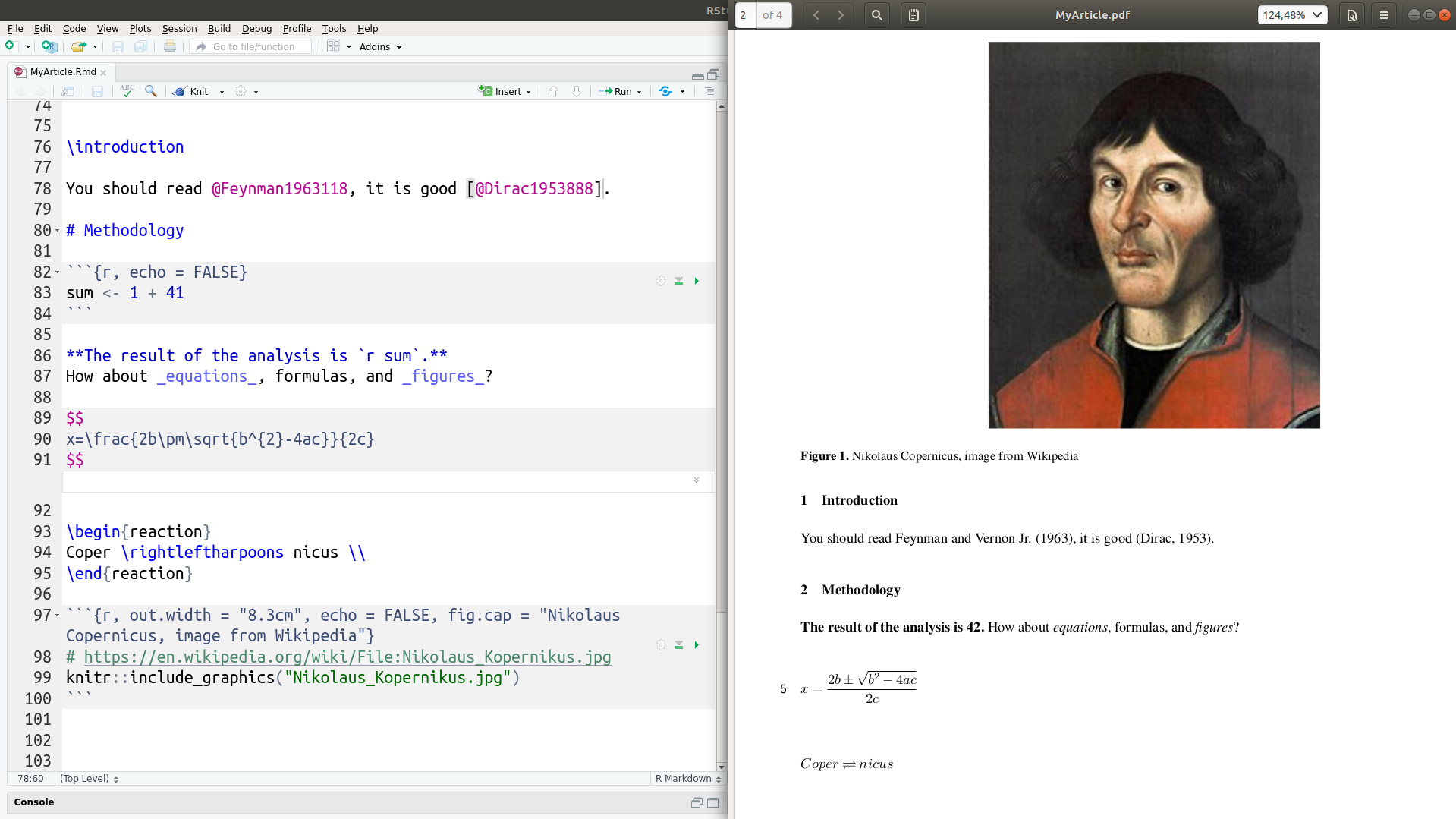
The commands created a directory with the Copernicus Publications template’s files, including an R Markdown (.Rmd) file ready to be edited by you (left-hand side of the screenshot), a LaTeX (.tex) file for submission to the publisher, and a .pdf file for inspecting the final results and sharing with your colleagues (right-hand side of the screenshot).
You can see how simple it is to format text, insert citations, chemical formulas or equations, and add figures, and how they are rendered into a high-quality output file.
All of these steps may also be completed with user-friendly forms when using RStudio, a popular development and authoring environment available for all operating systems. The left-hand side of the following screenshot shows the form for creating a new document based on a template, and the right-hand shows side the menu for rendering, called “knitting” with R Markdown because code and text are combined into one document like threads in a garment.
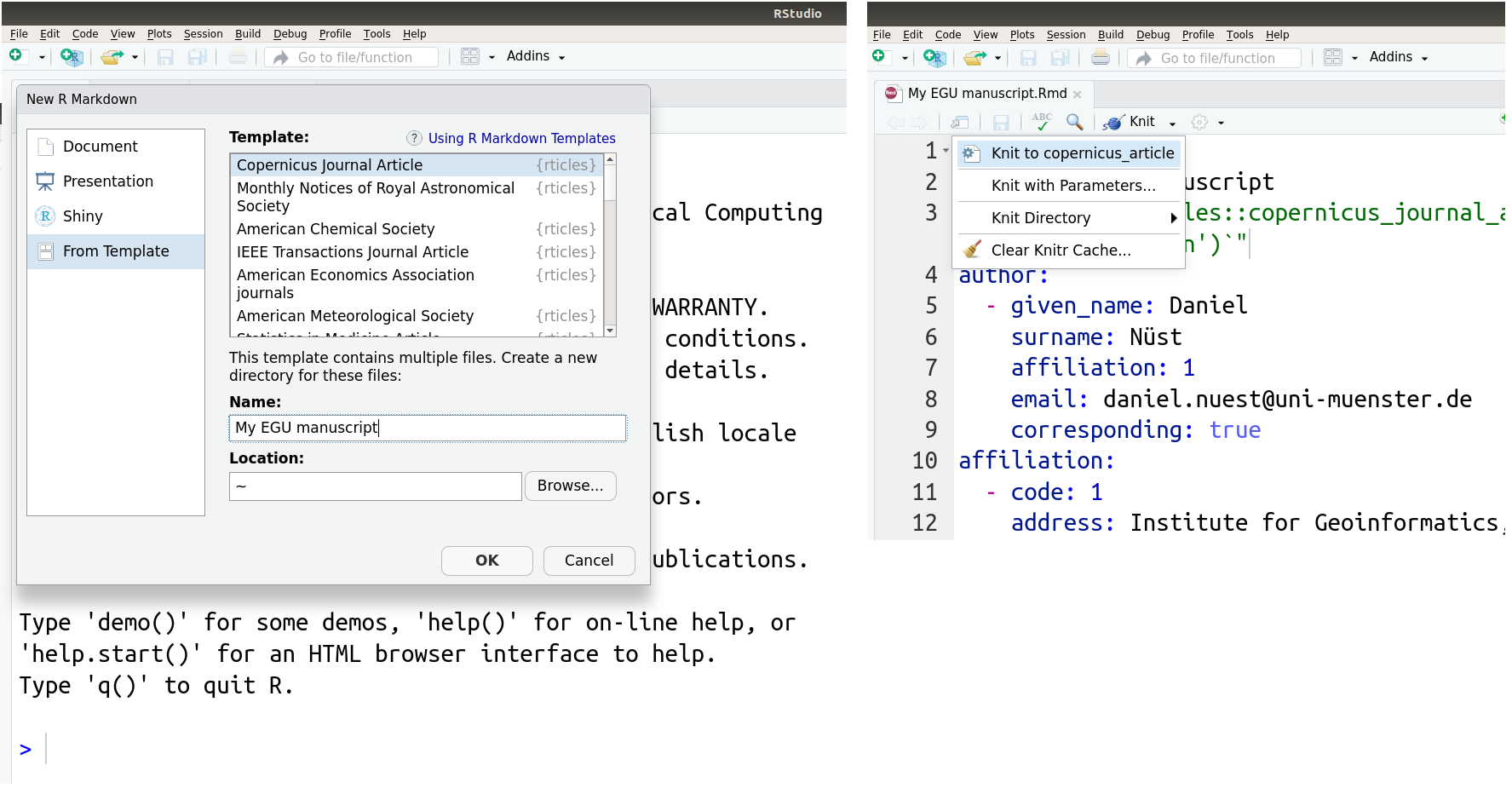
And in case you decide last minute to submit to a different journal, rticles supports many publishers so you only have to adjust the template while the whole content stays the same.
Sustainable access to supplemental data
Data published today should be published and properly cited using appropriate research data repositories following the FAIR data principles. Journals require authors to follow these principles, see for example the Copernicus Publications data policy or a recent announcement by Nature. Other publishers required, or still do today, to store supplemental information (SI), such as dataset files, extra figures, or extensive descriptions of experimental procedures, as part of the article. Usually only the article itself receives a digital object identifier (DOI) for long-term identification and availability. The DOI minted by the publisher is not suitable for direct access to supplemental files, because it points to a landing page about the identified object. This landing page is designed to be read by humans but not by computers.
The R package suppdata closes this gap.
It supports downloading supplemental information using the article’s DOI.
This way suppdata enables long-term reproducible data access when data was published as SI in the past or in exceptional cases today, for example if you write about a reproduction of a published article.
In the latest version available from GitHub (suppdata is on its way to CRAN) the supported publishers include Copernicus Publications.
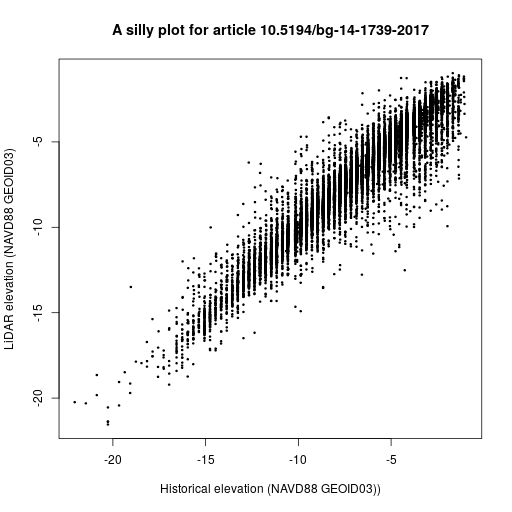
The following example code downloads a data file for the article “Divergence of seafloor elevation and sea level rise in coral reef ecosystems” by Yates et al. published in Biogeosciences in 2017.
The code then creates a mostly meaningless plot shown below.
# load required R extension package:
library("suppdata")
# download a specific supplemental information (SI) file
# for an article using the article's DOI:
csv_file <- suppdata::suppdata(
x = "10.5194/bg-14-1739-2017",
si = "Table S1 v2 UFK FOR_PUBLICATION.csv")
supplemental
# read the data and plot it (toy example!):
my_data <- read.csv(file = csv_file, skip = 3)
plot(x = my_data$NAVD88_G03, y = my_data$RASTERVALU,
xlab = "Historical elevation (NAVD88 GEOID03))",
ylab = "LiDAR elevation (NAVD88 GEOID03)",
main = "A data plot for article 10.5194/bg-14-1739-2017",
pch = 20, cex = 0.5)
Main takeaways
Authoring submission-ready manuscripts for journals of Copernicus Publications just got a lot easier. Everybody who can write manuscripts with a word processor can learn quickly R Markdown and benefit from a preproducible data science workflow. Digital notebooks not only improve day-to-day research habits, but the same workflow is suitable for authoring high-quality scholarly manuscripts and graphics. The interaction with the publisher is smooth thanks to the LaTeX submission format, but you never have to write any LaTeX. The workflow is based on an established Free and Open Source software stack and embraces the idea of preproducibility and the principles of Open Science. The software is maintained by an active, growing, and welcoming community of researchers and developers with a strong connection to the geospatial sciences. Because of the complete and consistent notebook, you, a colleague, or a student can easily pick up the work at a later time. The road to effective and transparent research begins with a first step - take it!
Acknowledgements
The software updates were contributed by Daniel Nüst from the project Opening Reproducible Research (o2r) at the Institute for Geoinformatics, University of Münster, Germany, but would not be able without the support of Copernicus Publications, the software maintainers most notably Yihui Xie and Will Pearse, and the general awesomeness of the R, R-spatial, Open Science, and Reproducible Research communities. The blog text was greatly improved with feedback by EGU’s Olivia Trani and Copernicus Publications’ Xenia van Edig. Thank you!
References
- Announcement: FAIR data in Earth science, Nature, 565(7738), 134–134, doi:10.1038/d41586-019-00075-3, 2019.
- Baker, M.: 1,500 Scientists Lift the Lid on Reproducibility, Nature, 533(7604), 452–454, doi:10.1038/533452a, 2016.
- Barba, L. A.: Terminologies for Reproducible Research, ArXiv:1802.03311 [Cs], February 9, 2018.
- Fanelli, D.: Opinion: Is Science Really Facing a Reproducibility Crisis, and Do We Need It To?, Proceedings of the National Academy of Sciences, 115(11), 2628–2631, doi:10.1073/pnas.1708272114, 2018.
- Gil, Y., David, C. H., Demir, I., Essawy, B. T., Fulweiler, R. W., Goodall, J. L., Karlstrom, L., Lee, H., Mills, H. J., Oh, J.-H., Pierce, S. A., Pope, A., Tzeng, M. W., Villamizar, S. R. and Yu, X.: Towards the Geoscience Paper of the Future: Best Practices for Documenting and Sharing Research from Data to Software to Provenance, Earth and Space Science, 3(10), 388–415, doi:10.1002/2015ea000136, 2016.
- Hardwicke, T. E., Mathur, M. B., MacDonald, K., Nilsonne, G., Banks, G. C., Kidwell, M. C., Hofelich Mohr, A., Clayton, E., Yoon, E. J., Henry Tessler, M., Lenne, R. L., Altman, S., Long, B. and Frank, M. C.: Data availability, reusability, and analytic reproducibility: evaluating the impact of a mandatory open data policy at the journal Cognition, Royal Society Open Science, 5(8), 180448, doi:10.1098/rsos.180448, 2018.
- Konkol, M. and Kray, C.: In-depth examination of spatiotemporal figures in open reproducible research, Cartography and Geographic Information Science, 1–16, doi:10.1080/15230406.2018.1512421, 2018.
- Konkol, M., Kray, C. and Pfeiffer, M.: Computational reproducibility in geoscientific papers: Insights from a series of studies with geoscientists and a reproduction study, International Journal of Geographical Information Science, 1–22, doi:10.1080/13658816.2018.1508687, 2018.
- Markowetz, F.: Five selfish reasons to work reproducibly, Genome Biology, 16(1), doi:10.1186/s13059-015-0850-7, 2015.
- Marwick, B., Boettiger, C. and Mullen, L.: Packaging Data Analytical Work Reproducibly Using R (and Friends), The American Statistician, 72(1), 80–88, doi:10.1080/00031305.2017.1375986, 2017.
- Munafò, M. R., Nosek, B. A., Bishop, D. V. M., Button, K. S., Chambers, C. D., Percie du Sert, N., Simonsohn, U., Wagenmakers, E.-J., Ware, J. J. and Ioannidis, J. P. A.: A manifesto for reproducible science, Nature Human Behaviour, 1(1), 21, doi:10.1038/s41562-016-0021, 2017.
- Nüst, D., Granell, C., Hofer, B., Konkol, M., Ostermann, F. O., Sileryte, R. and Cerutti, V.: Reproducible research and GIScience: an evaluation using AGILE conference papers, PeerJ, 6, e5072, doi:10.7717/peerj.5072, 2018.
- Ostermann, F. O. and Granell, C.: Advancing Science with VGI: Reproducibility and Replicability of Recent Studies using VGI, Transactions in GIS, 21(2), 224–237, doi:10.1111/tgis.12195, 2016.
- Pearse, W. D. and A Chamberlain, S.: Suppdata: Downloading Supplementary Data from Published Manuscripts, Journal of Open Source Software, 3(25), 721, doi:10.21105/joss.00721, 2018.
- ReproZip: Computational Reproducibility With Ease, F. Chirigati, R. Rampin, D. Shasha, and J. Freire. In Proceedings of the 2016 ACM SIGMOD International Conference on Management of Data (SIGMOD), pp. 2085-2088, 2016
- Sandve, G. K., Nekrutenko, A., Taylor, J. and Hovig, E.: Ten Simple Rules for Reproducible Computational Research, edited by P. E. Bourne, PLoS Computational Biology, 9(10), e1003285, doi:10.1371/journal.pcbi.1003285, 2013.
- Stark, P. B.: Before reproducibility must come preproducibility, Nature, 557(7707), 613–613, doi:10.1038/d41586-018-05256-0, 2018.
- Toelch, U. and Ostwald, D.: Digital open science—Teaching digital tools for reproducible and transparent research, PLOS Biology, 16(7), e2006022, doi:10.1371/journal.pbio.2006022, 2018.
- Jupyter, P., Bussonnier, M., Forde, J., Freeman, J., Granger, B., Head, T., Holdgraf, C., Kelley, K., Nalvarte, G., Osheroff, A., Pacer, M., Panda, Y., Perez, F., Ragan-Kelley, B. and Willing, C.: Binder 2.0 - Reproducible, interactive, sharable environments for science at scale, in Proceedings of the 17th Python in Science Conference, SciPy., 2018.
- Wilson, G., Bryan, J., Cranston, K., Kitzes, J., Nederbragt, L. and Teal, T. K.: Good enough practices in scientific computing, PLOS Computational Biology, 13(6), e1005510, doi:10.1371/journal.pcbi.1005510, 2017.
- Yates, K. K., Zawada, D. G., Smiley, N. A. and Tiling-Range, G.: Divergence of seafloor elevation and sea level rise in coral reef ecosystems, Biogeosciences, 14(6), 1739–1772, doi:10.5194/bg-14-1739-2017, 2017.


