Two new publications and one survey in September 2023
18 Sep 2023 | By Eftychia KoukourakiWe are excited to share the latest conference presentations of past O2R team members! Even though the project now delivered, the dedication of the people who were instrumental in our now-delivered project still contributes to the field.
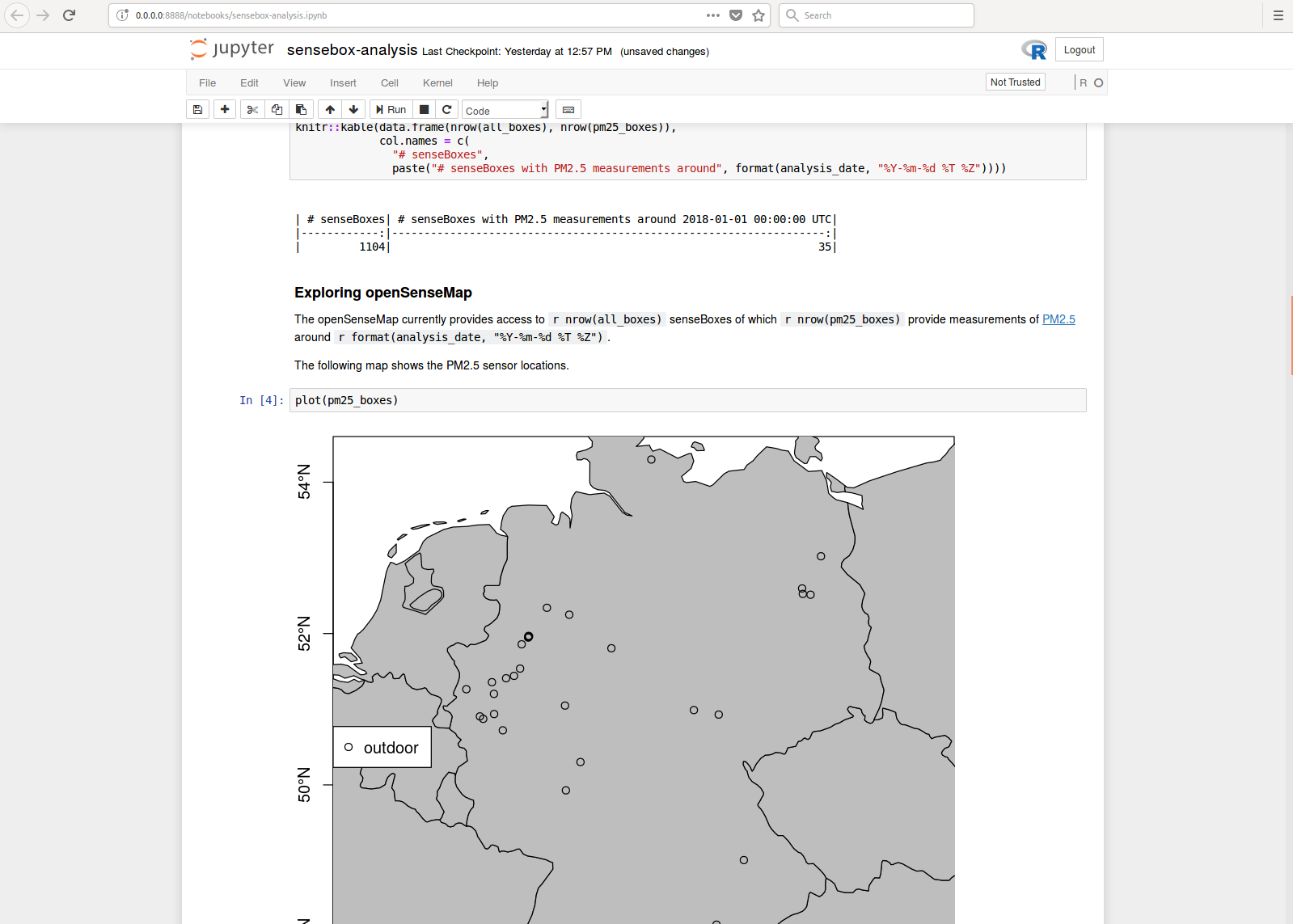
On September 6th, Edzer Pebesma presented his work Reproducing Spatial Data Science Publications at the 4th Spatial Data Science Symposium, where he discusses the challenges posed by the always evolving software, the several dependencies and the publishing industry, reports on experiences from developer communities, and looks at the convergence in the spatial data science software ecosystems.
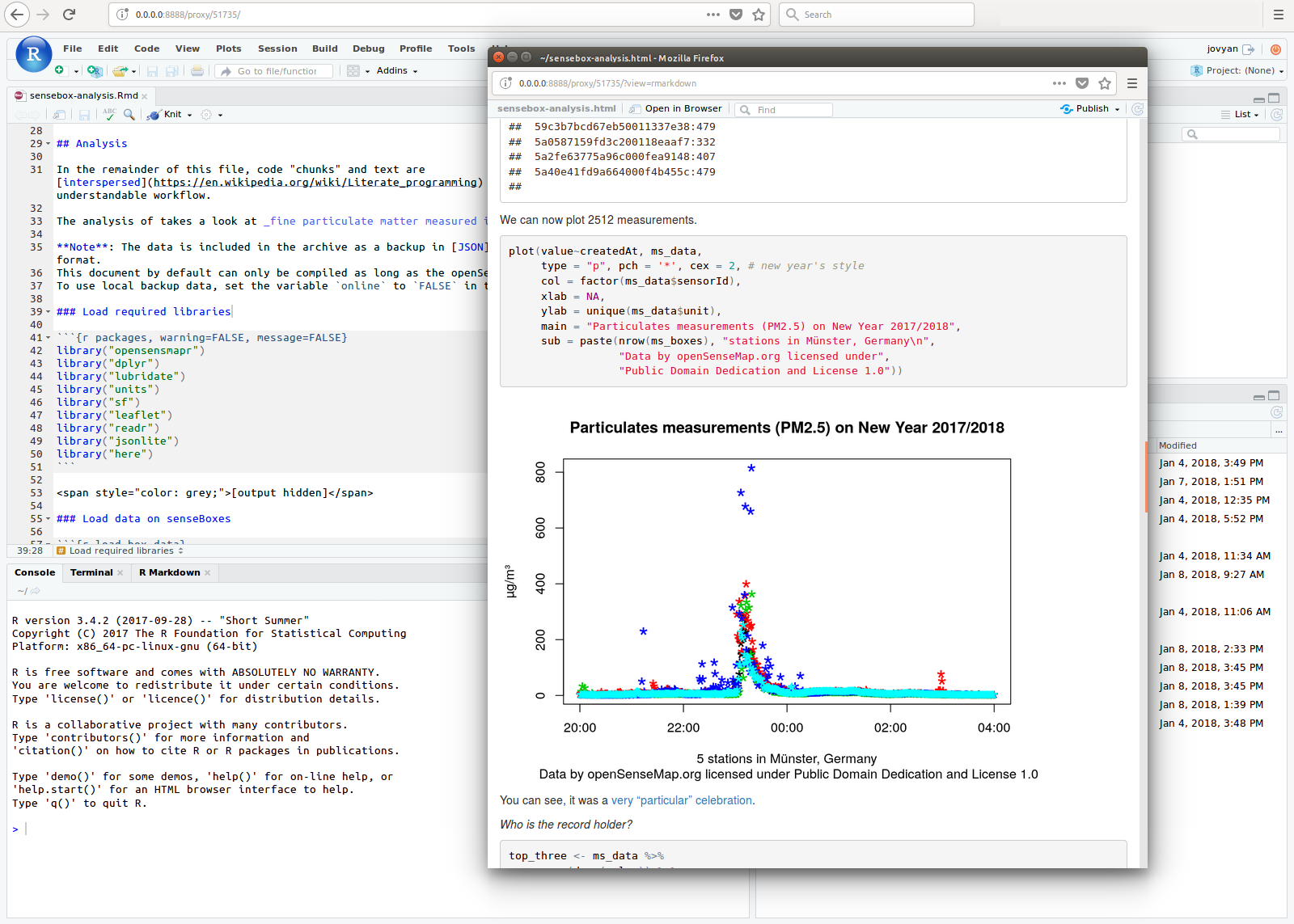
On September 13th, I presented the exploratory study that we elaborated with Christian Kray about Map Reproducibility in Geoscientific Publications at the 12th International Conference on Geographic Information Science (GIScience 2023), where we suggest an inceptive set of criteria to assess the success of map reproduction and a set of guidelines for improving map reproducibility in geoscientific publications.
If you want to help us understand better the notion of map reproducibility by sharing your POV, take part in the survey that is already up and running: https://t.ly/qquoj.




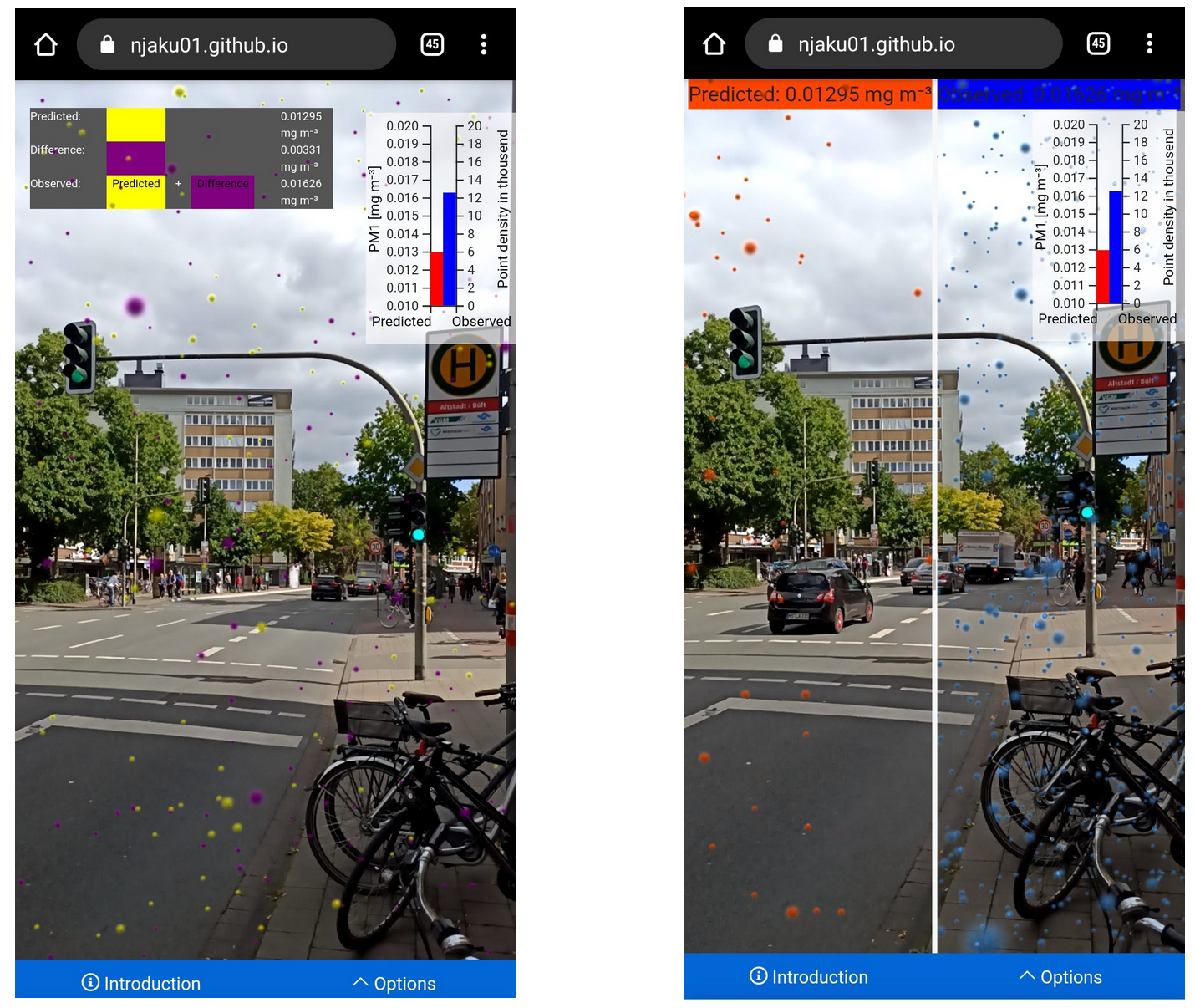



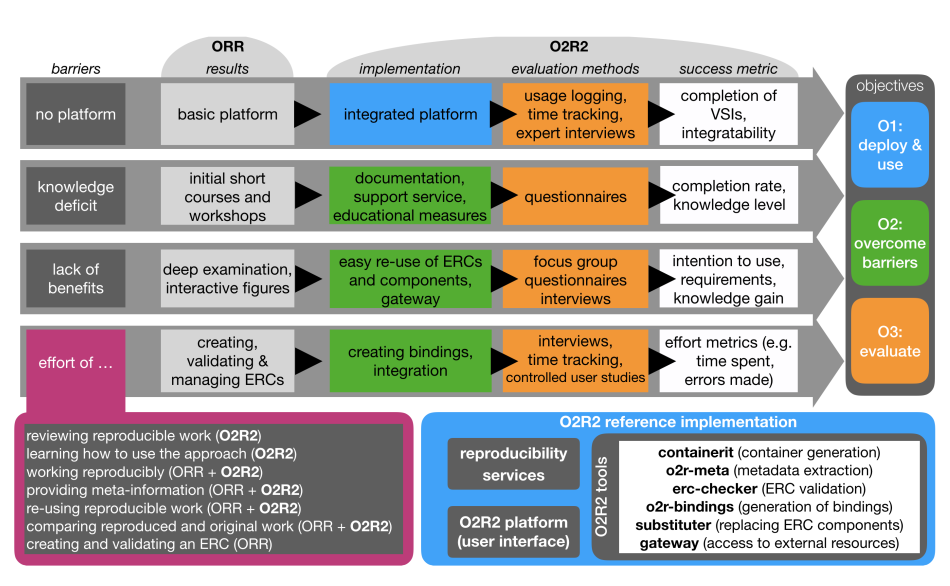

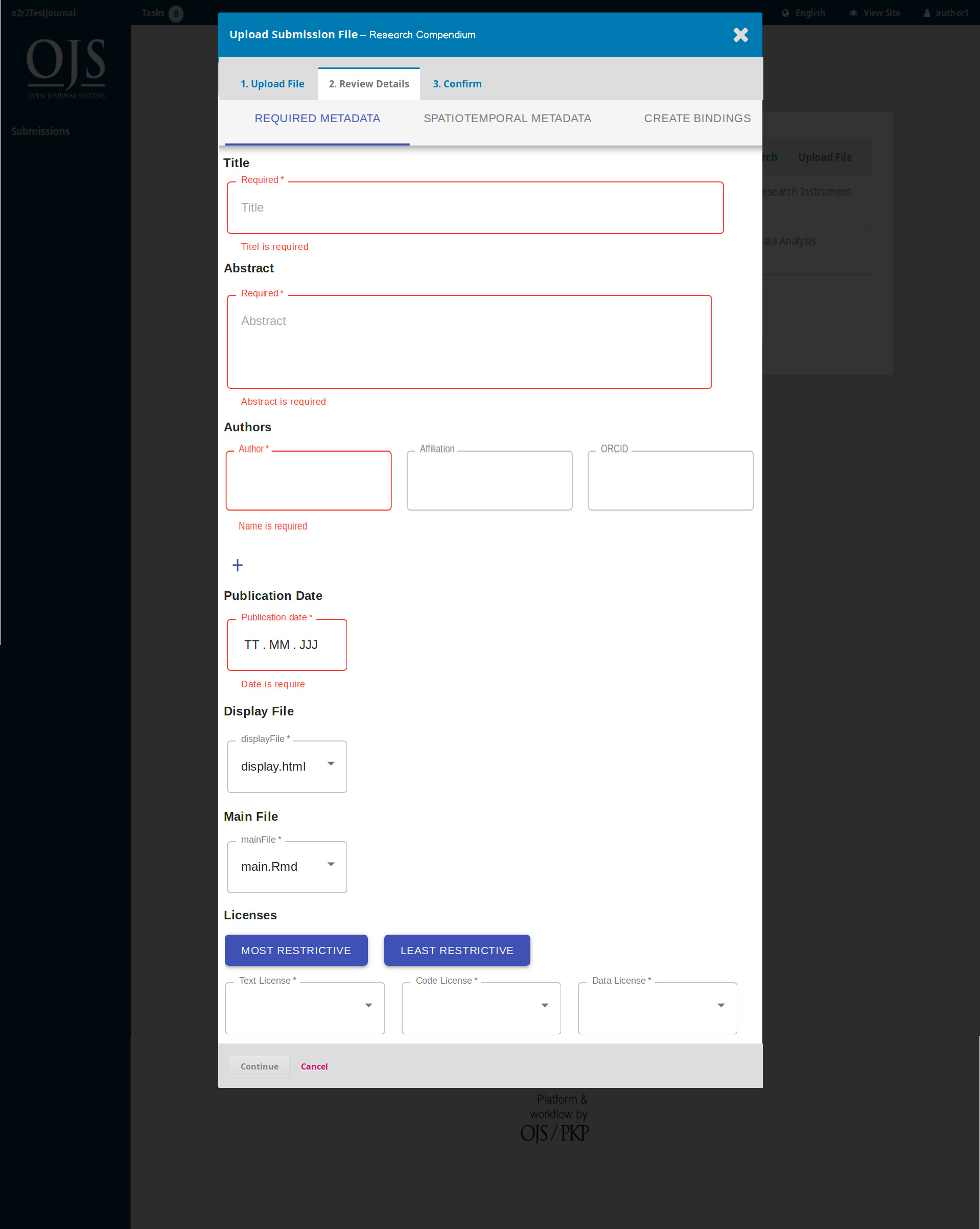
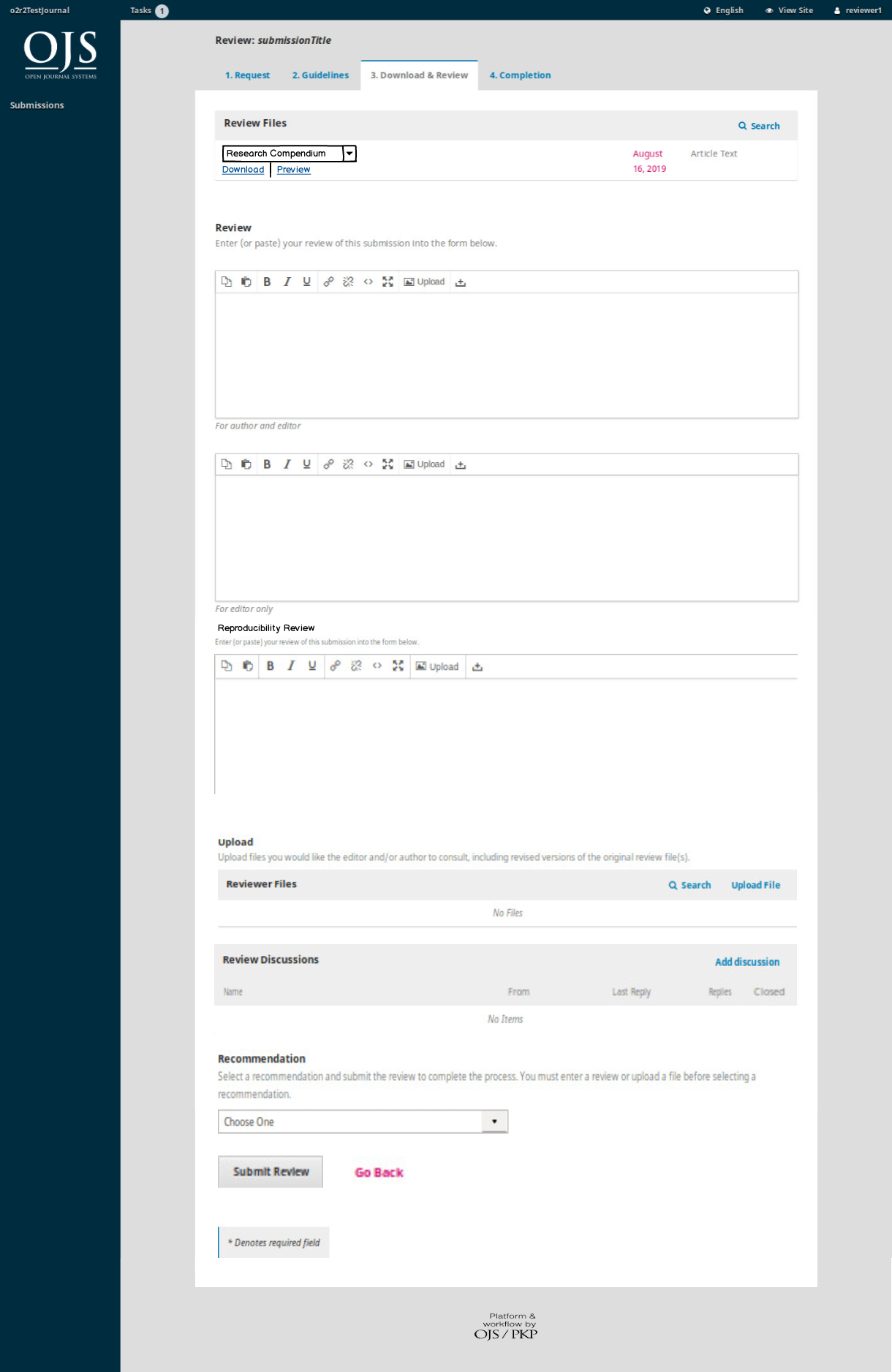
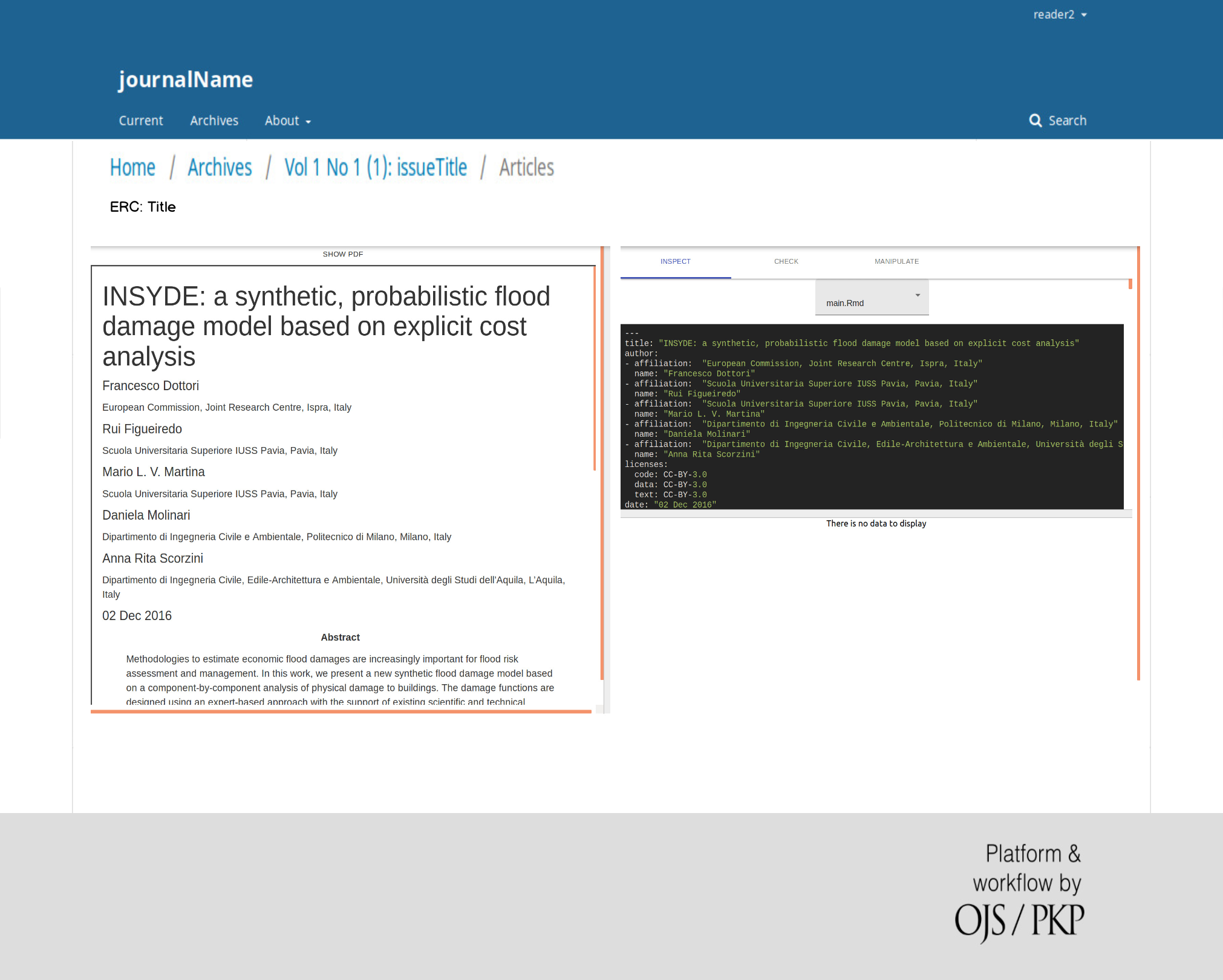
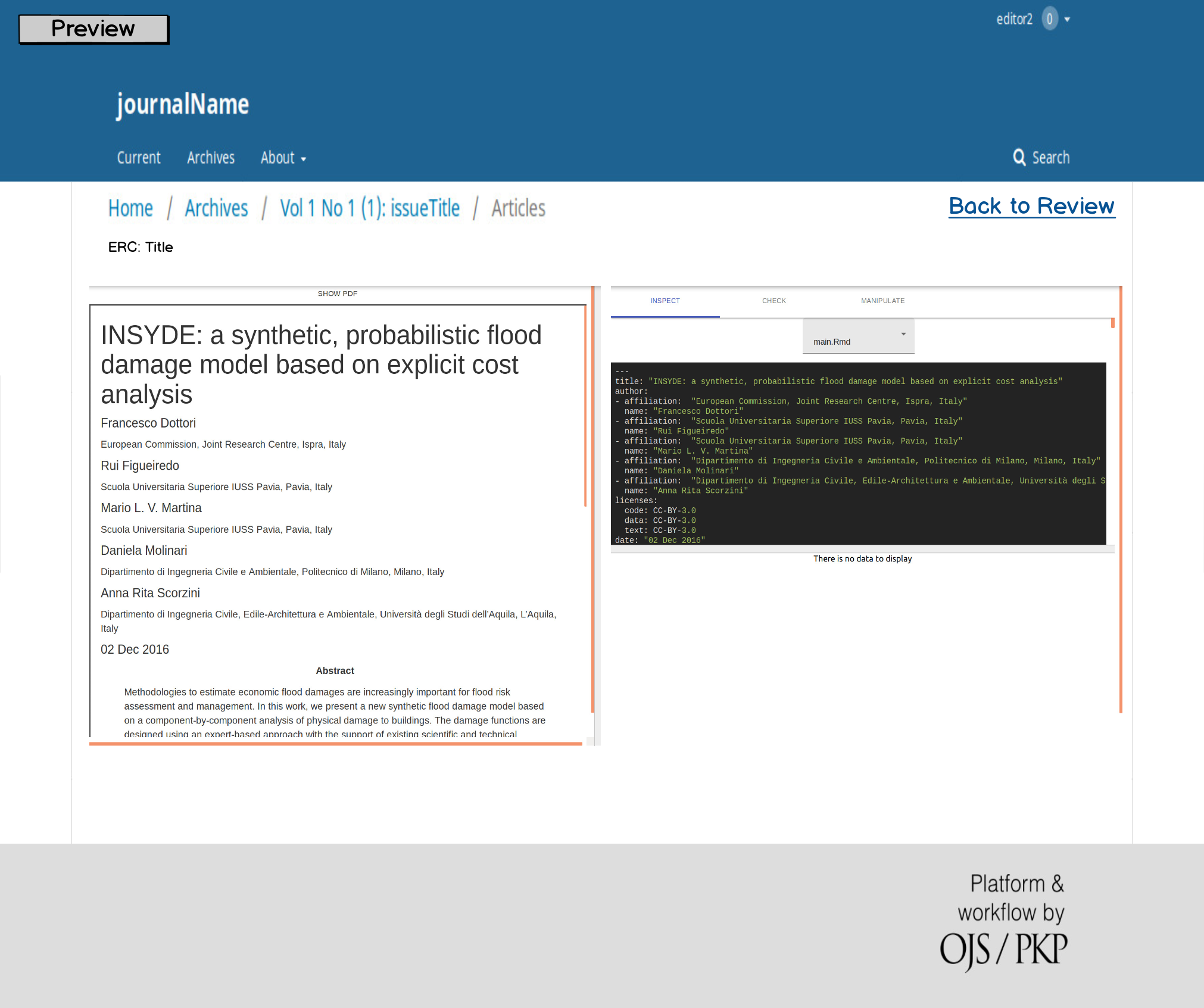
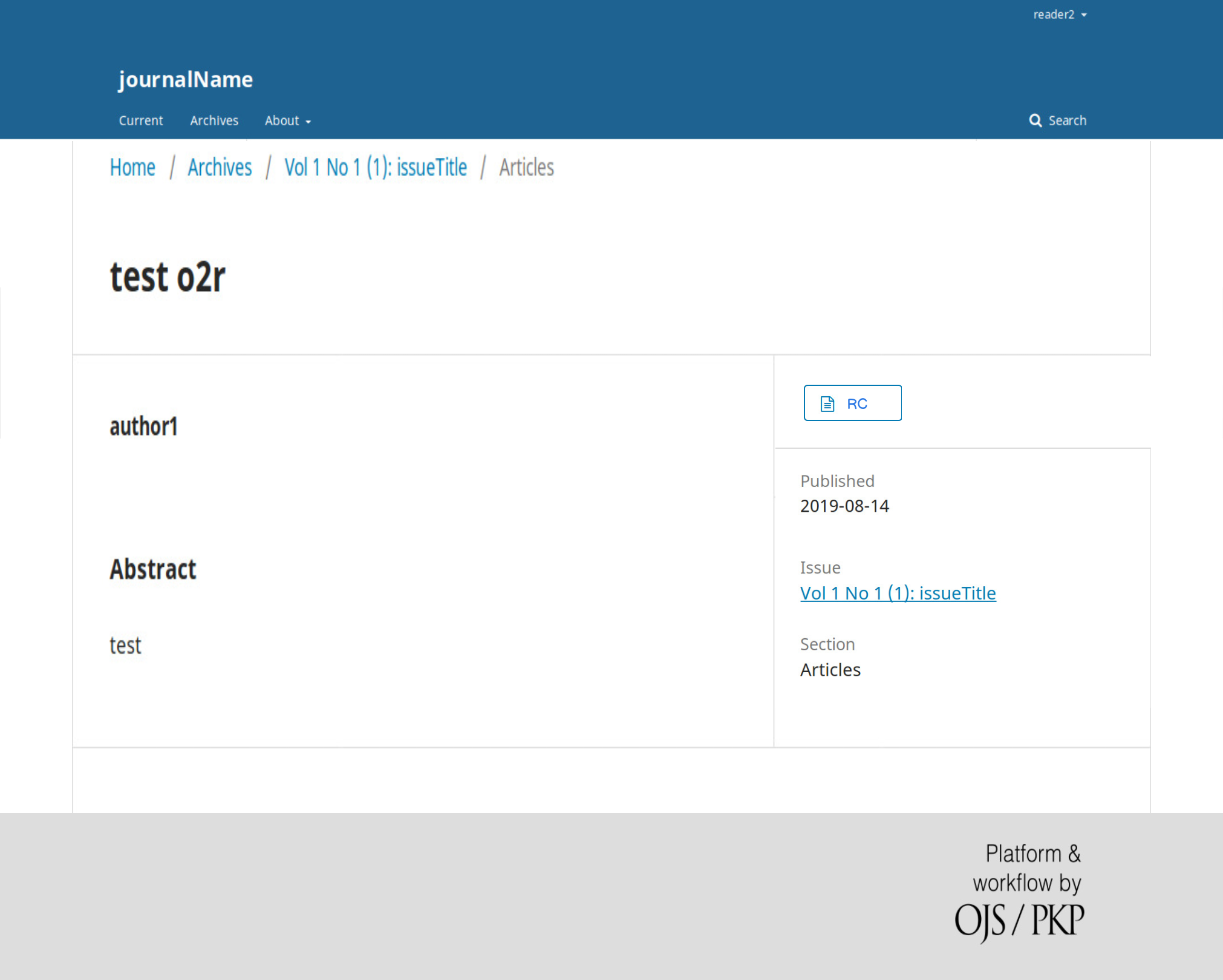
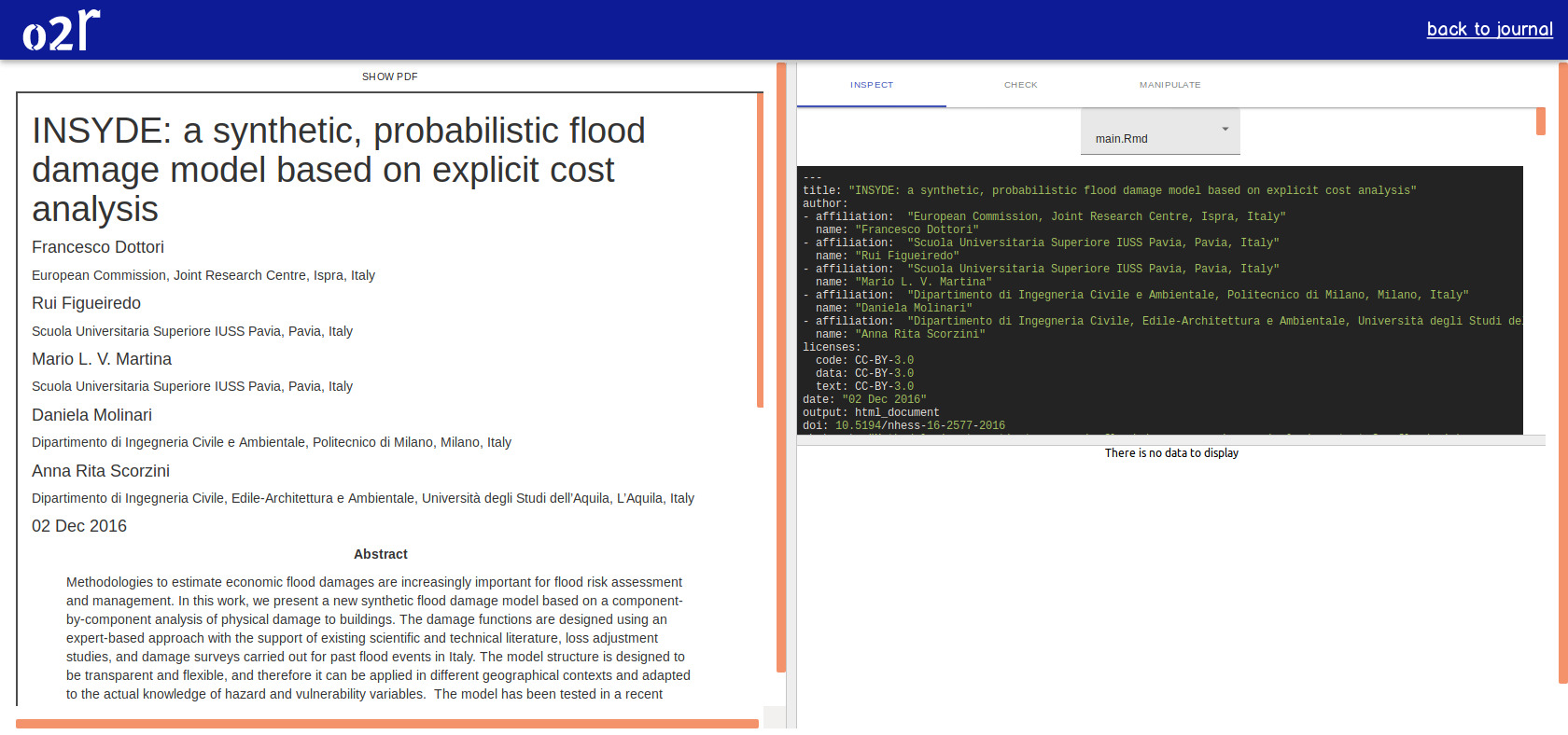
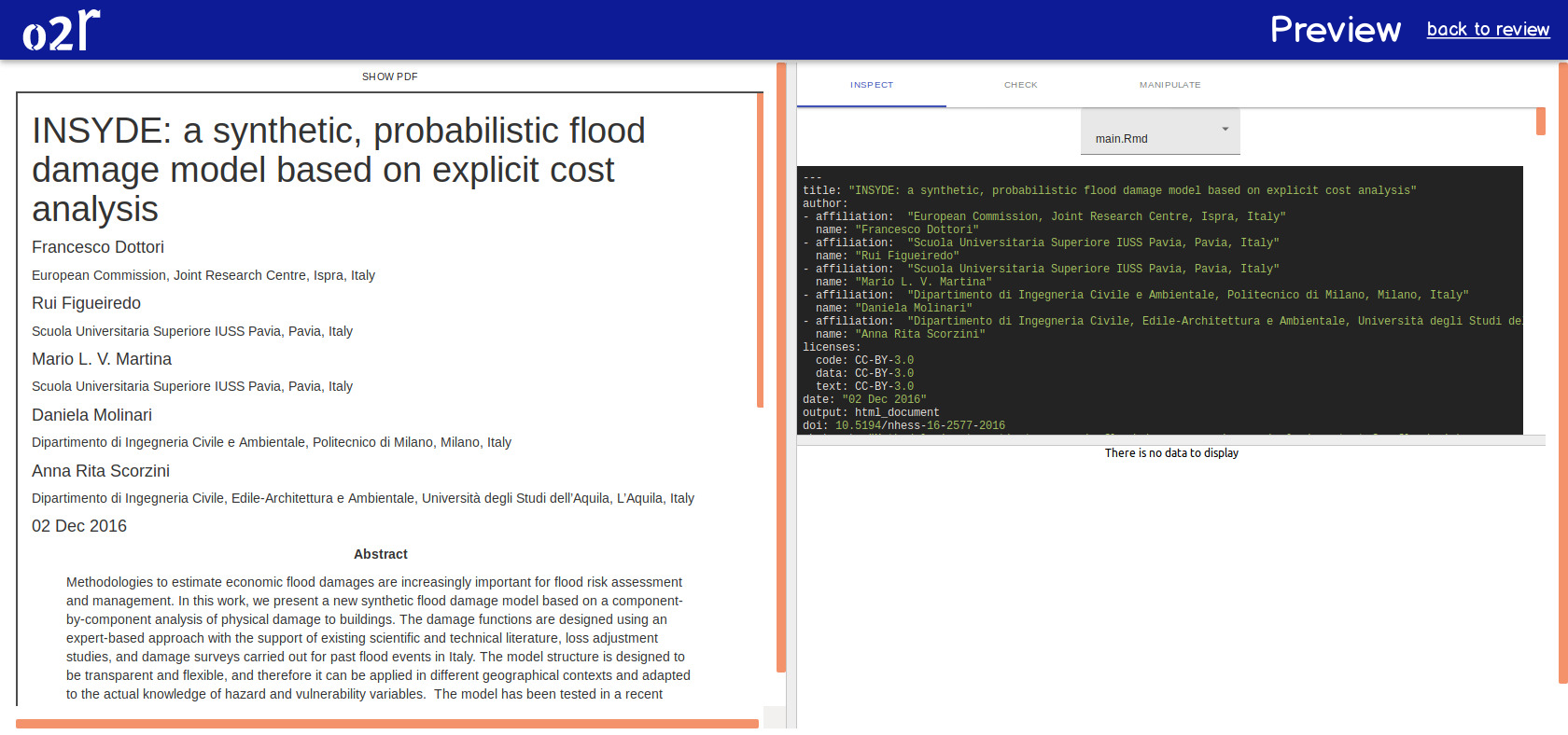


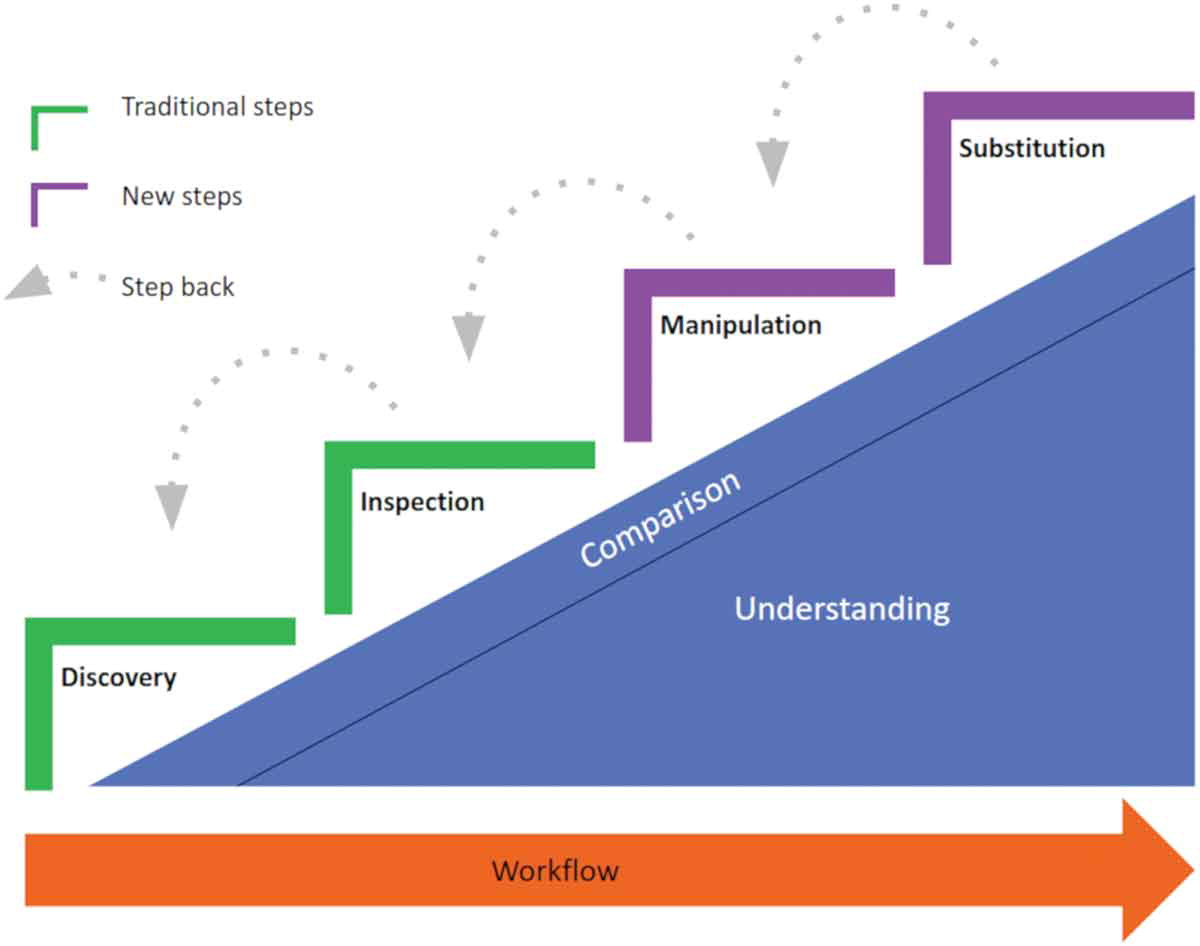

 On April 1st 2019 the o2r team started into a new phase (“o2r2”).
In the next 30 months we plan to put our prototypes to the test with real articles, of course not without considerably improving them beforehand.
On April 1st 2019 the o2r team started into a new phase (“o2r2”).
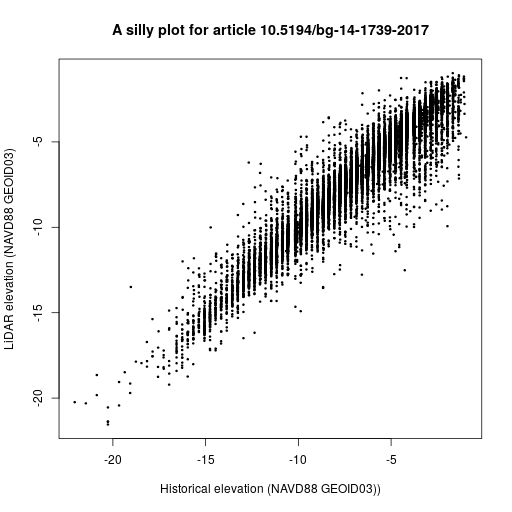
In the next 30 months we plan to put our prototypes to the test with real articles, of course not without considerably improving them beforehand.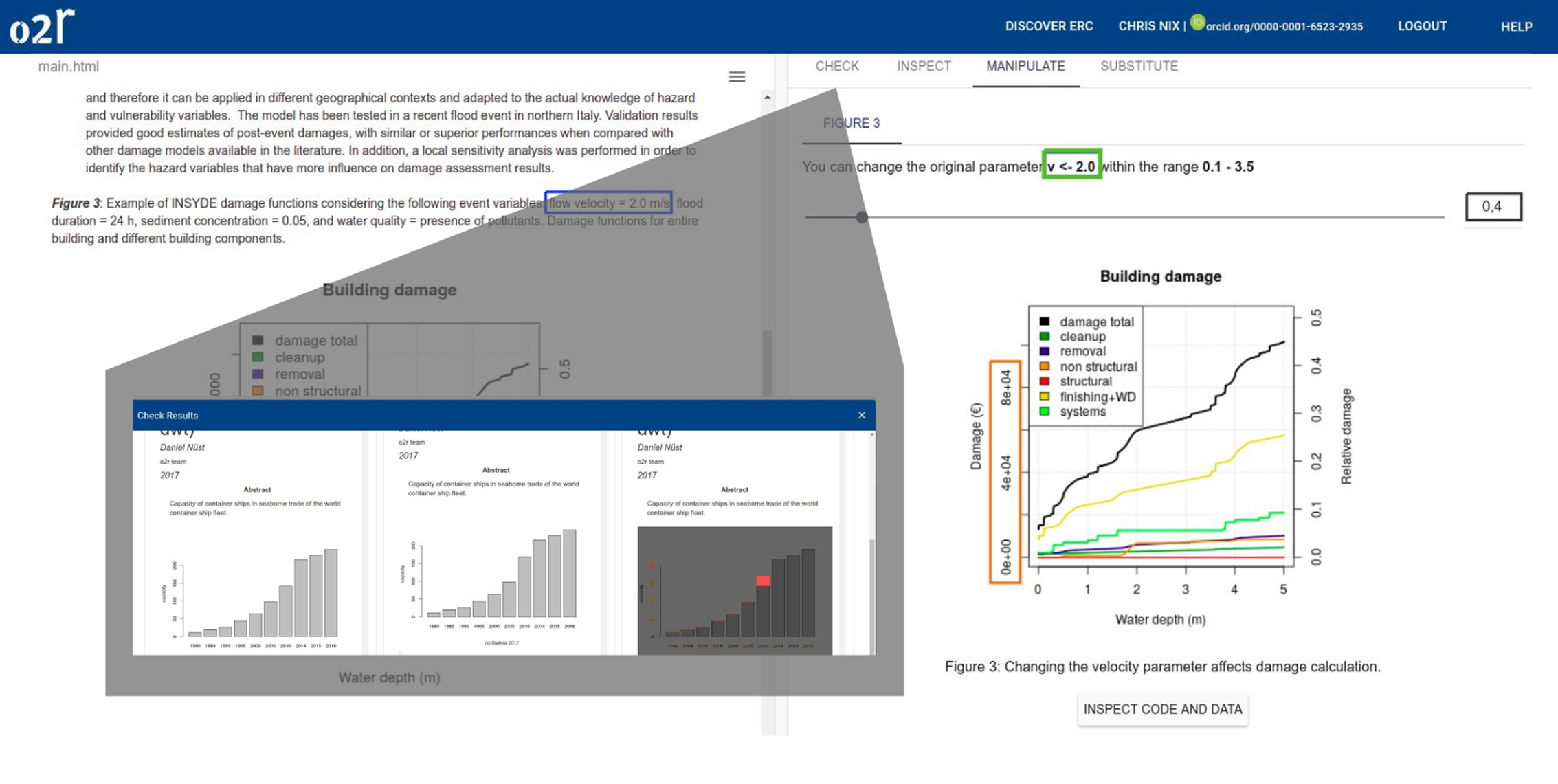



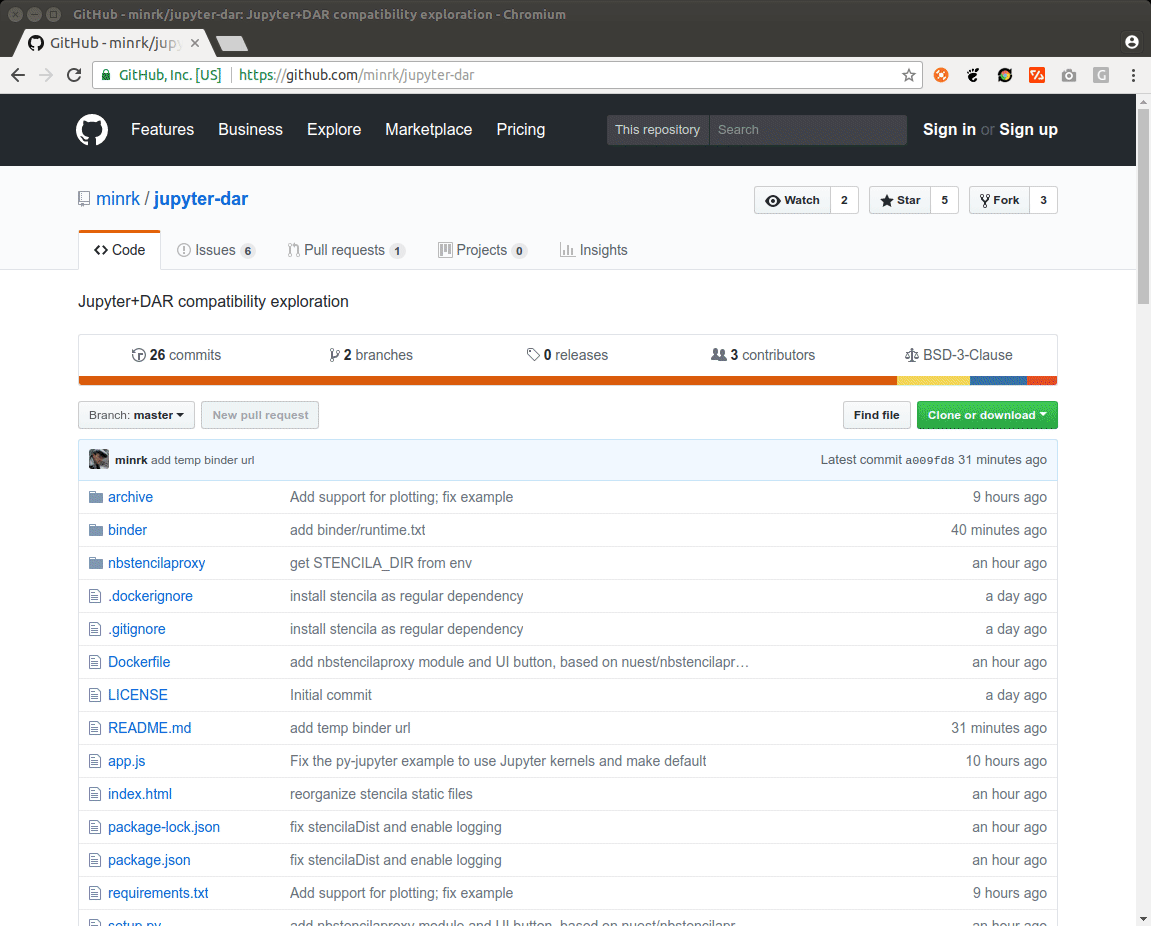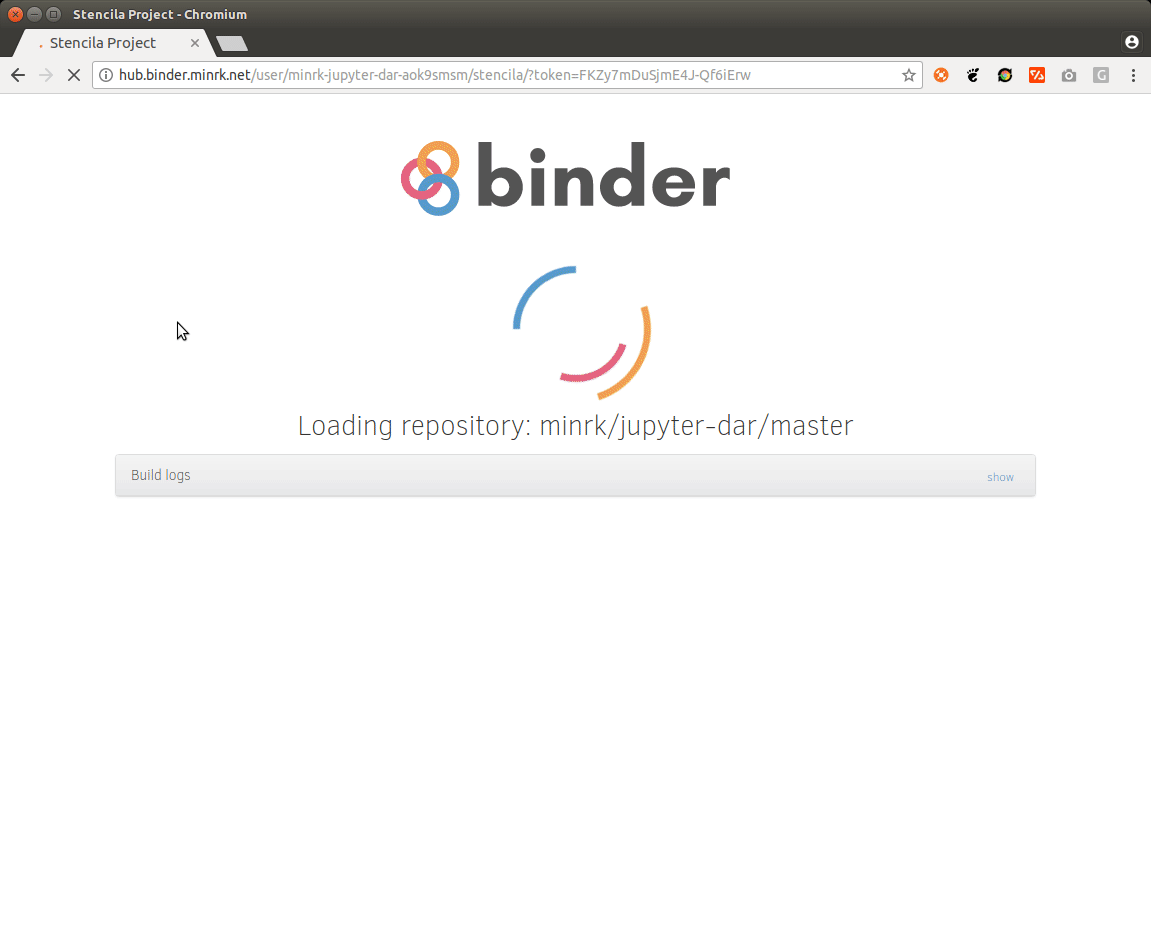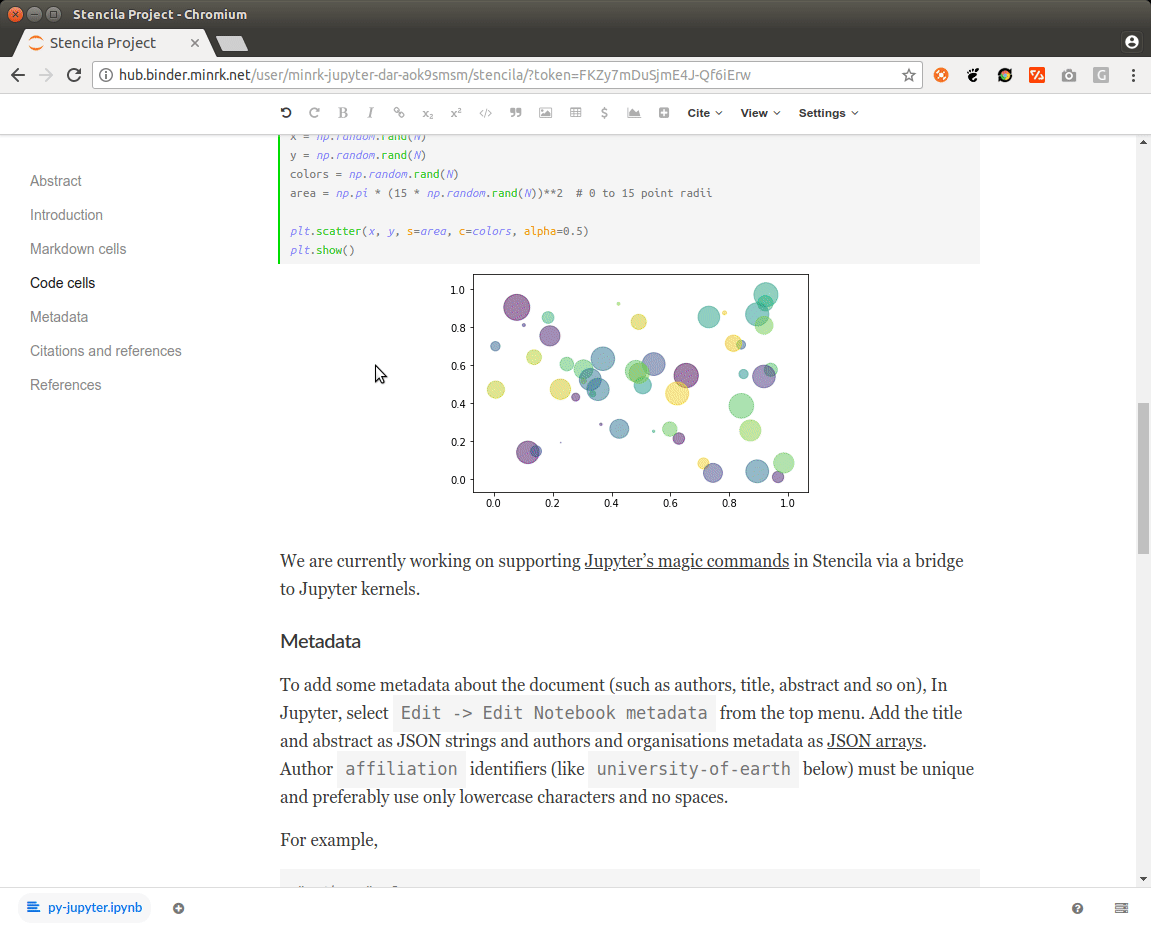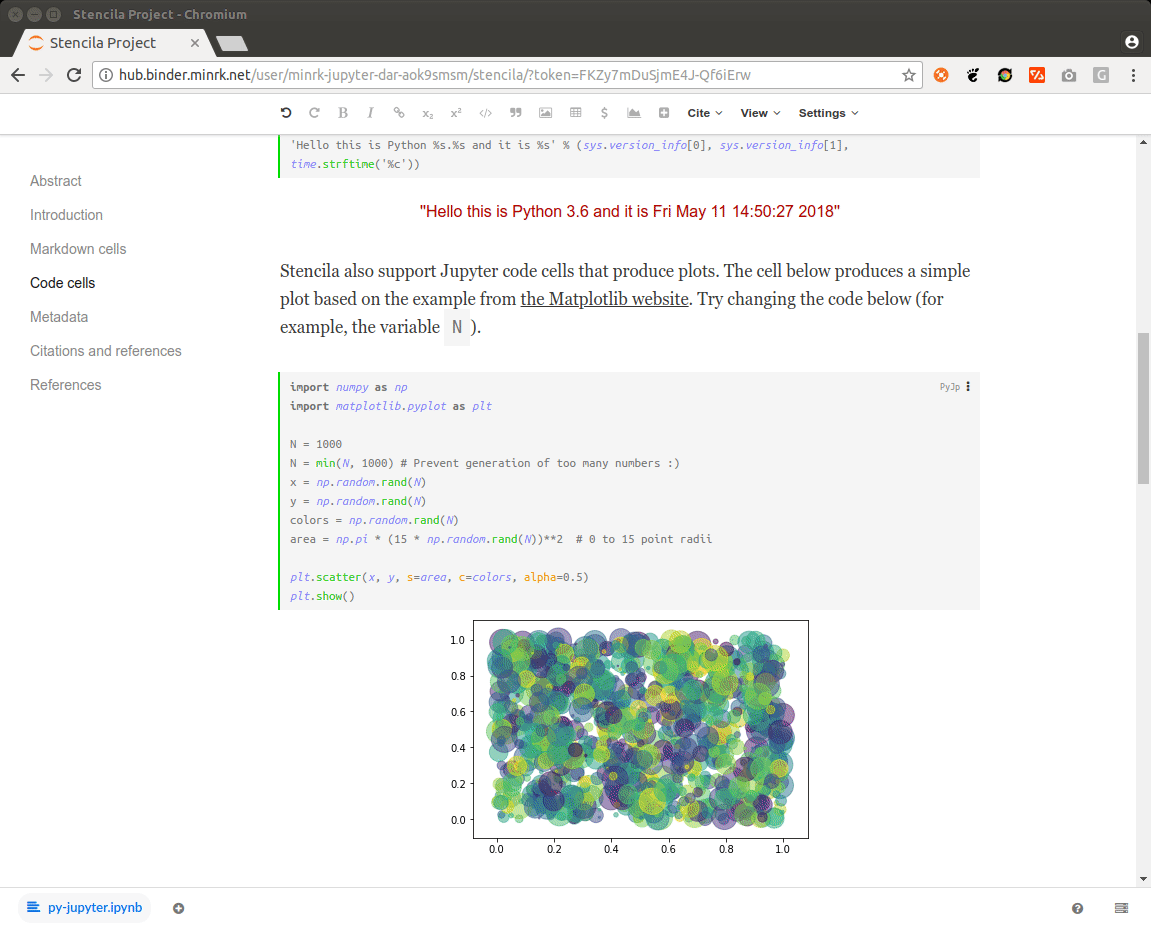 [
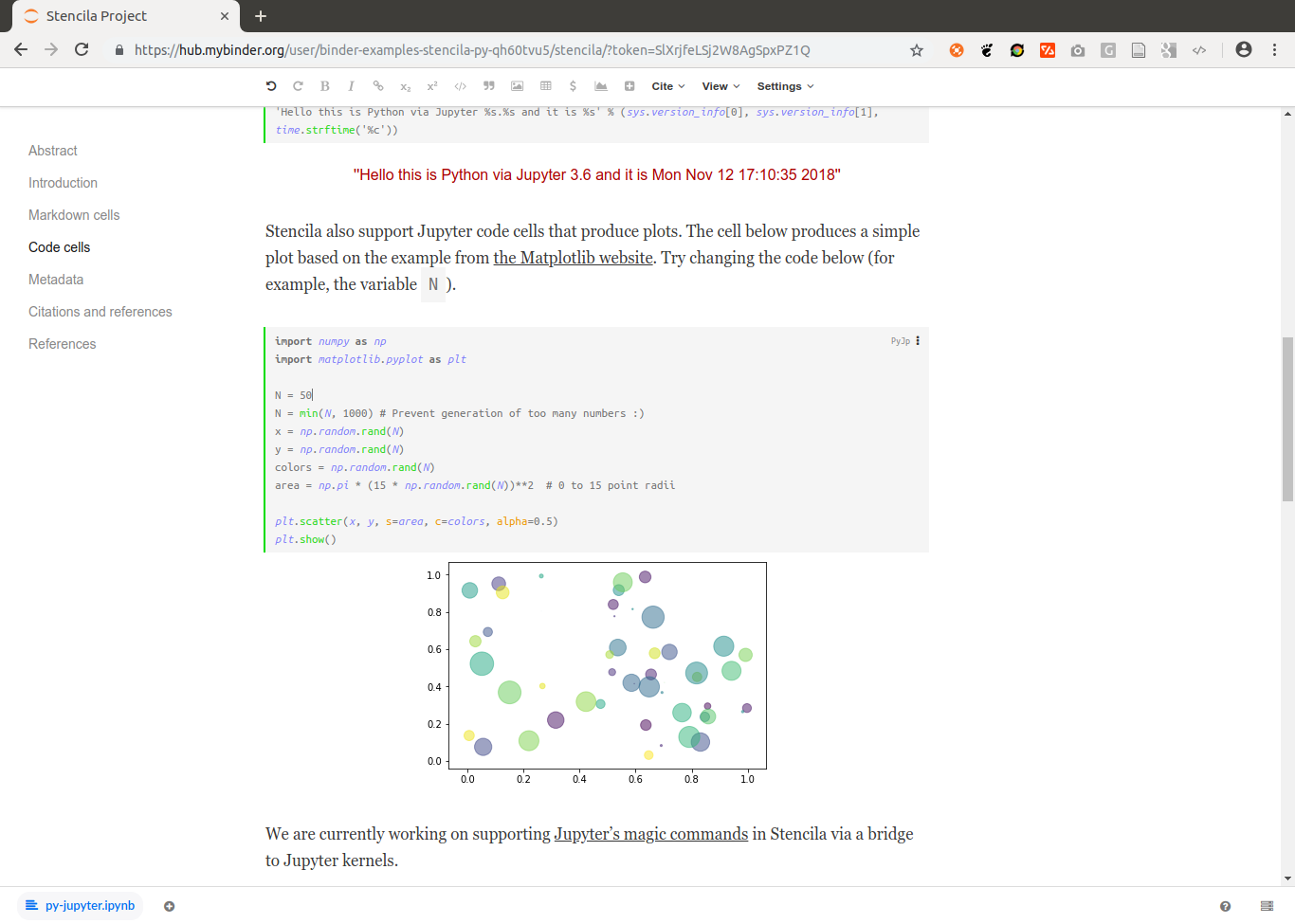
[
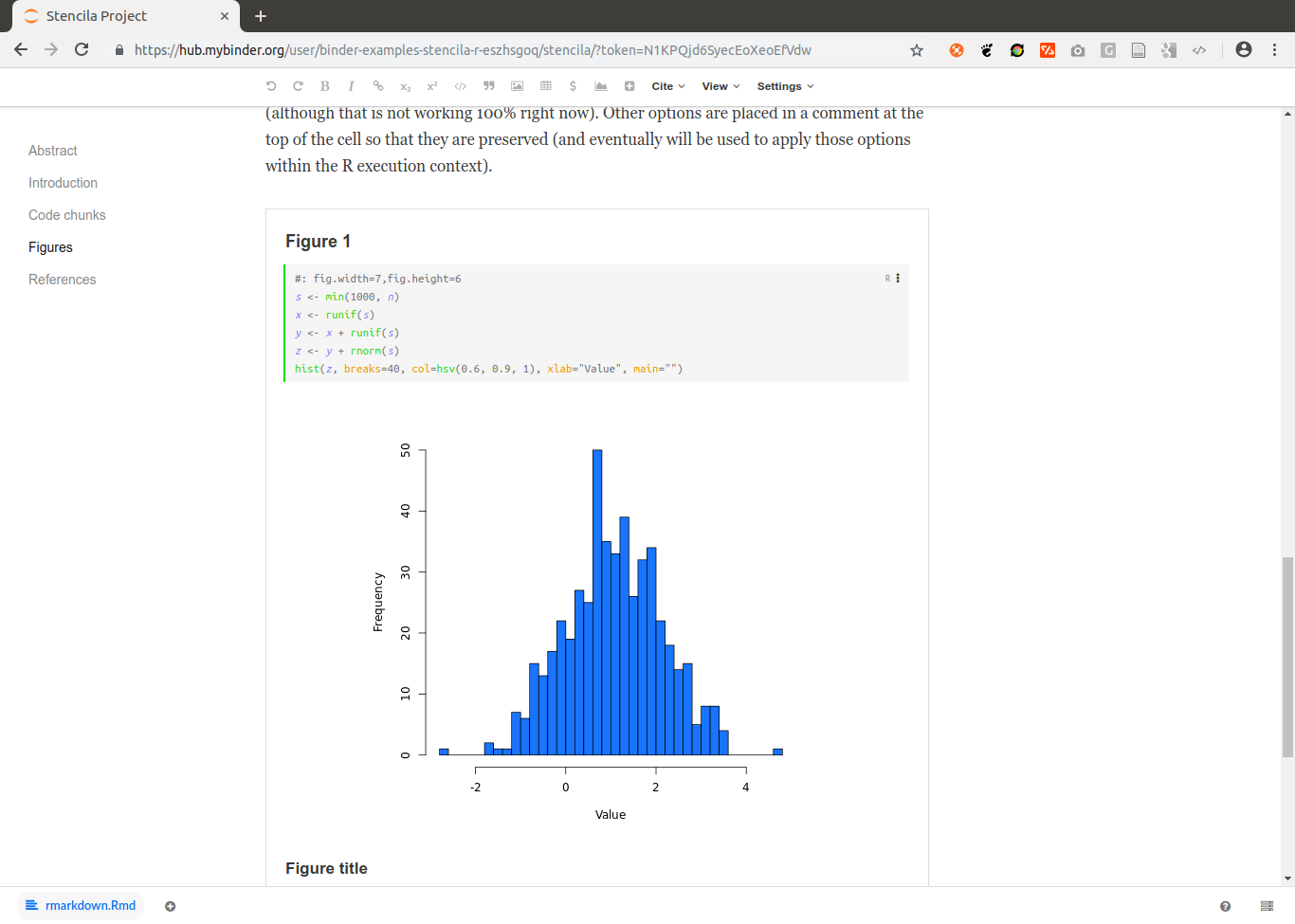












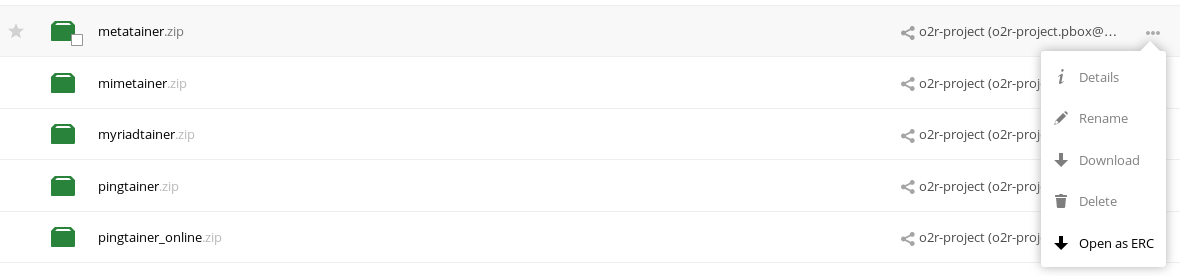
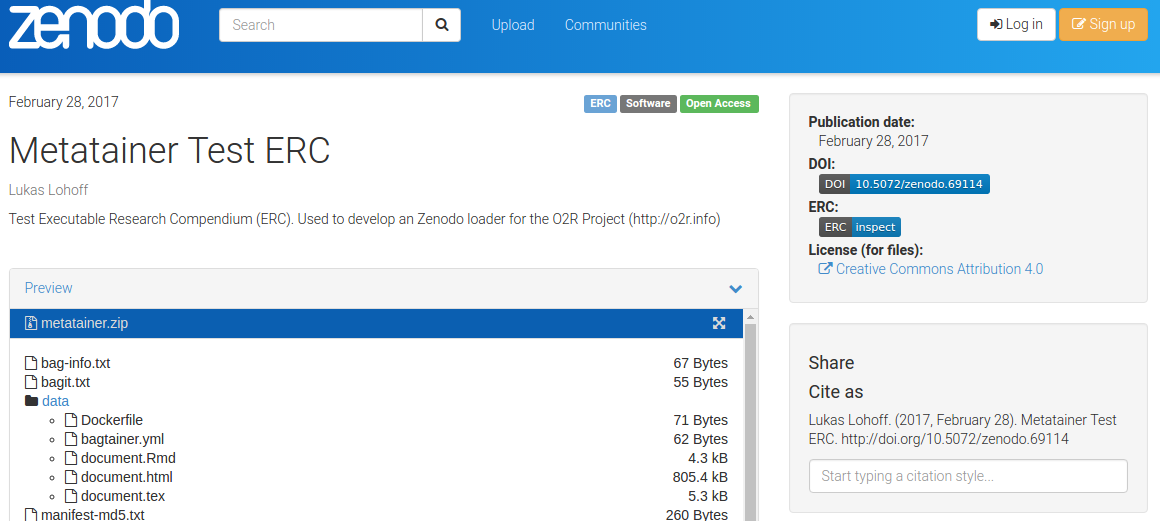













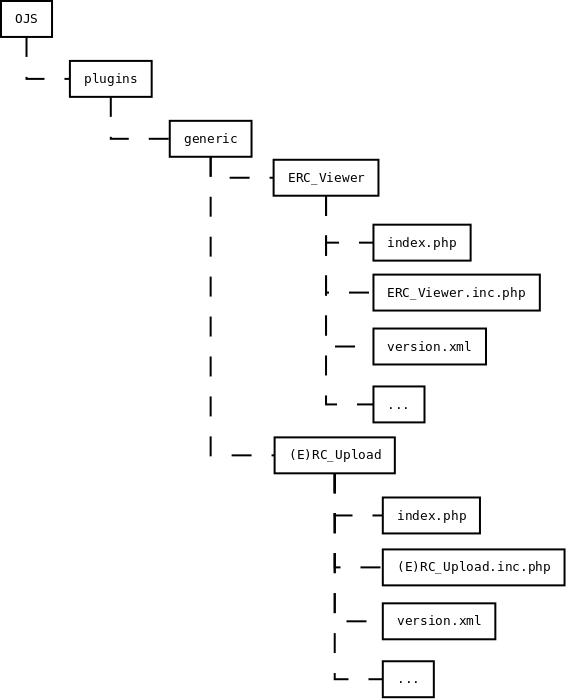
 As some popular projects surprisingly did not have Docker images yet, Daniel started a new independent project
As some popular projects surprisingly did not have Docker images yet, Daniel started a new independent project 


 Next week the largest European geosciences conference of the year will take place in Vienna: the
Next week the largest European geosciences conference of the year will take place in Vienna: the 







